
























Detecting Microbiological Colonies in Food Samples
FSD | 3M Health Care (Solventum) | 2018 - 2021
View my associated patent here: Microorganic Detection System with Worklists
3M’s Food Safety Department (FSD) was relaunching their Petrifilm Plater Reader device and wanted to develop a companion software platform to differentiate it from the competition and create a superior customer experience. While food safety is integral to our everyday modern lives, technology adoption in the industry is behind the curve with many labs still enumerating colonies by hand and documenting and storing test results in paper-based binders or in on-premise LIMs systems. Our team of two was asked to develop an approachable, yet powerful software platform that increased productivity and supported food safety labs and technicians in multiple aspects of their roles such as colony enumeration, documentation and traceability, reporting and trend analysis.
Challenge
Phase One: Iterative Learning Loops
I was fortunate to have the opportunity to work with a team of multi-disciplinary experts from the FSD division, including microbiologists, engineers, product owner, developers, and marketing professionals who had been working for a year before I joined the project to identify the required imaging technology (i.e. camera and lens) and begin the development of the colony enumeration algorithms.
This meant the team was ready to dive into exploration of software features and functionality as well as UI development. My design colleague and I had inherited initial wireframes and research from a previous UX designer assigned to the project and quickly began to develop a testing plan to continue the design process.
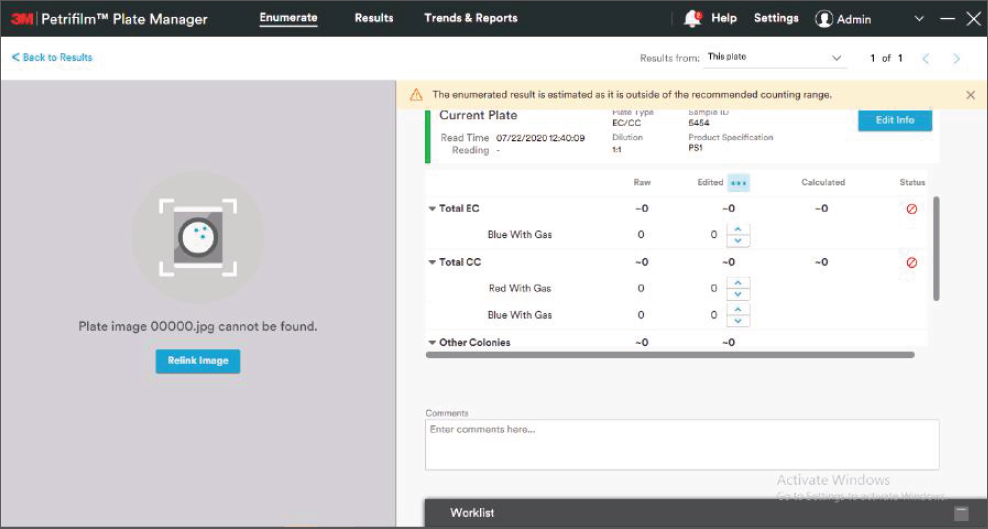
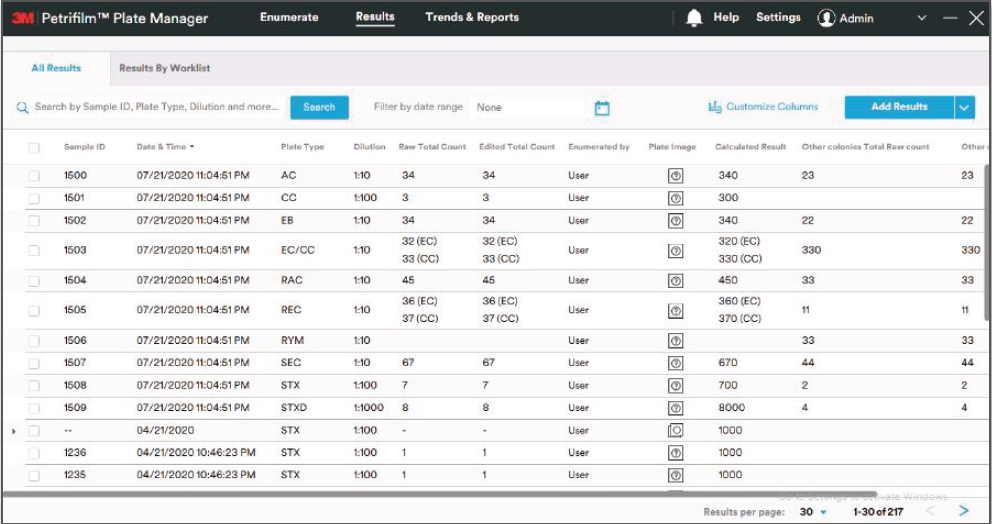
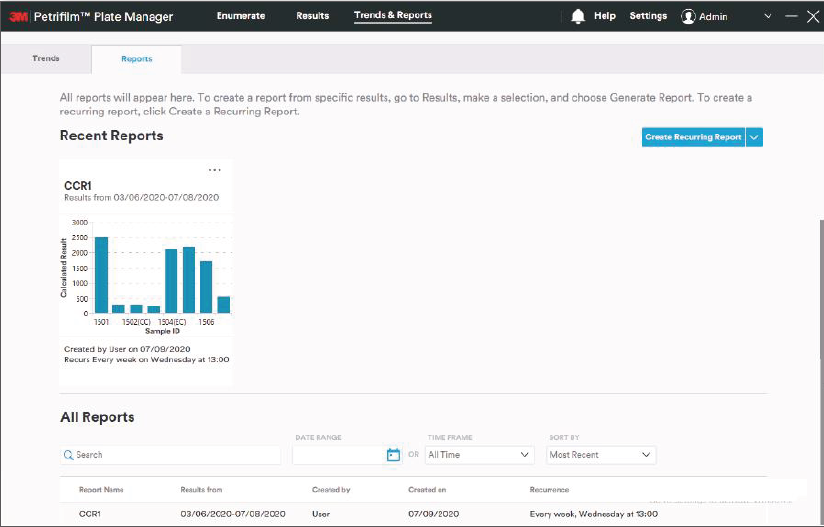
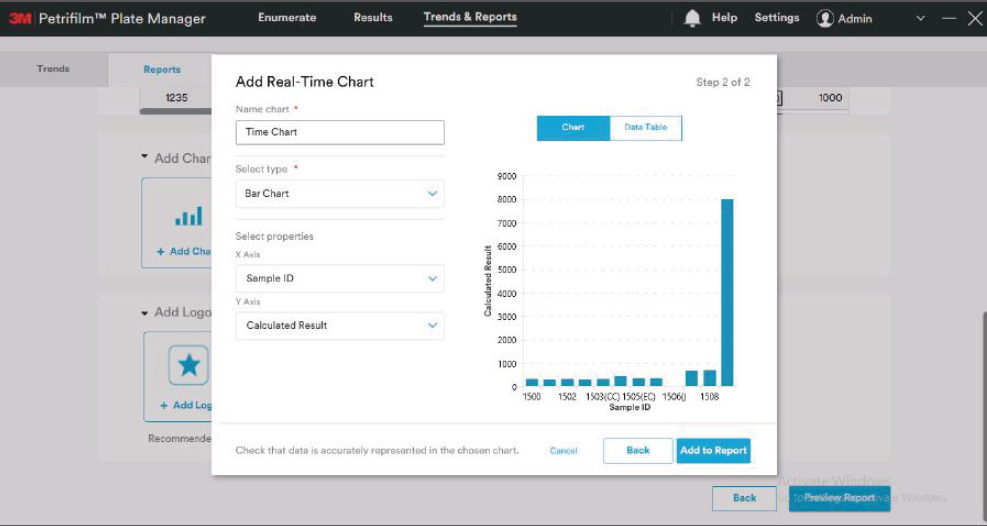
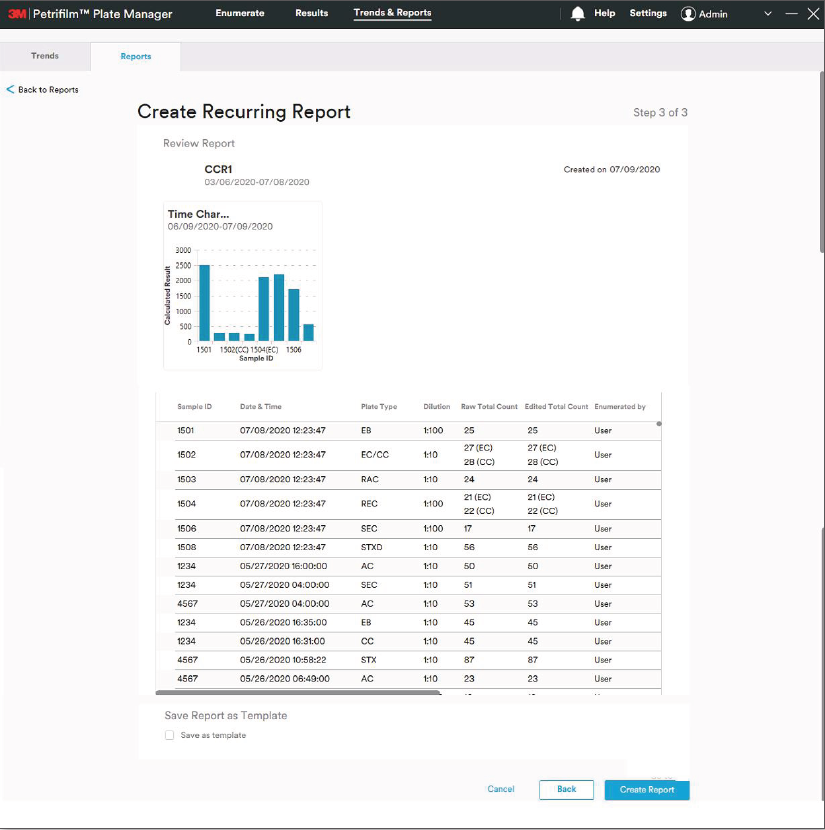
We first took a pass at defining an initial information architecture, grouping the many functions of the software underneath a simple three-step structure: Enumerate, Review and Report. We then developed clickable prototypes for each of the software “tabs” to identify data and interaction requirements.
We then conducted a series of design sprints, each sprint focusing on a different function, and quickly refined our prototypes and designs between each test. I took the lead on the development of the research protocols, and creation of the clickable prototypes while my colleague focused on the visual design of the UI.
Outcomes
Validation of the proposed information architecture of the software platform
Discovery and documentation of user requirements for the different software functions - both technical and experience - for different user types
Development of clickable, easy-to-iterate software prototypes for continued testing and validation
Refined software visual style and UI design
Phase Two: Design and Development
As the overall features and functionality began to fall into place, my teammate and I turned our attention to the more detailed, science-specific, and hardware-related components of the Petrifilm Plate Manager Software.
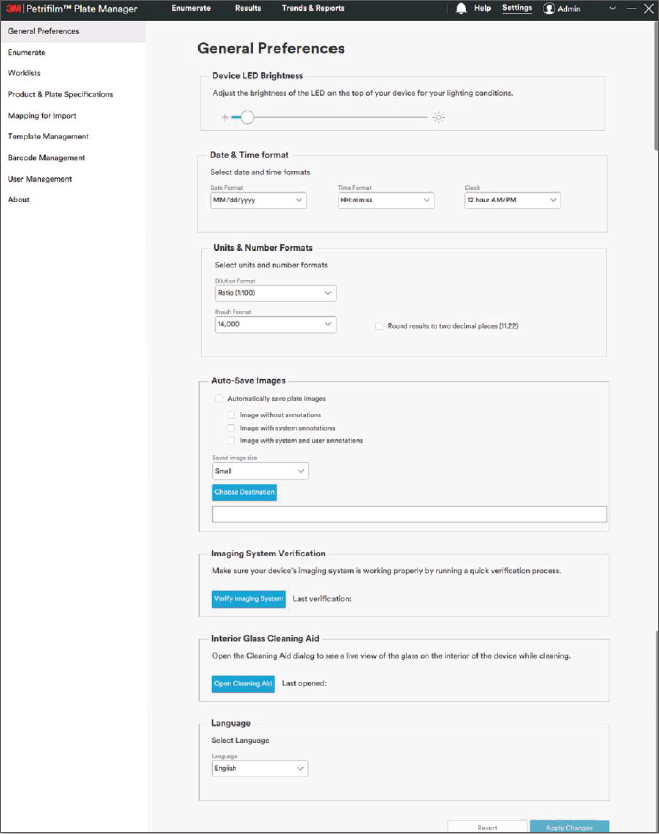
I was responsible for defining the backend utilities and the ‘settings’ specifications needed to be included as well as driving the interaction design for these components. As PPRA is a global product it had to include a variety of customizable settings for different food safety standards, testing protocols, reporting regulations, units of measure, and even different workflows depending on which country or region a customer was located in. We spent time with 3M’s global tech services team members - who were mostly trained microbiologists themselves - to gather the requirements and best practices for the different regions. I then worked with our development team to design a flexible import and export function for test specifications, customer preferences, barcode symbologies, and global standards, among others.
My colleague was focused on developing a visual language and icon library for the enumeration function of the software- defining the symbols, colors, and differentiating visual features for each of the 10 different Petrifilm Plate types. This amounted a a massive library with well over 100 specific icons, each signifying a specific function, edit, or colony type.
Outcomes
Developed a library of user stories and associated user flow for launch and future releases
Designed and integrated sound and haptic feedback into the user experience to aid usability when taking photos of wounds in inconvenient locations, the first feature to do so in the 3M Health Care digital portfolio
Launched R1 of the feature into the existing My Wound Healing app in Q1 of 2022.

